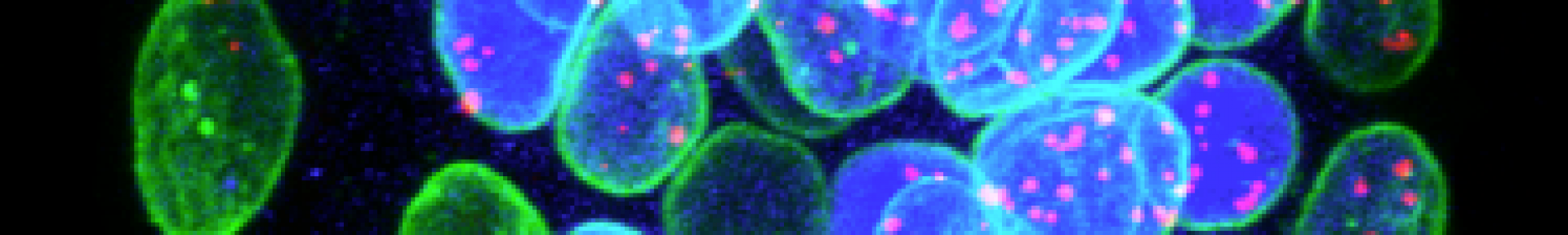
The epiblast cells will produce all the cells of the future individual and its descendants. Moreover, the epi is the source of the famous ES pluripotent stem cells (Nobel Prize in 2007 by Evans, Smithies, Capecchi) or similar to iPS reprogrammed cells (Nobel Prize in 2012 by Yamanaka). These cells have the capacity to give any embryonic or adult cell type and therefore have a great potential for cell therapy.
Our team is studying how in the embryo Epi cells acquire these "pluripotent" properties and how they differentiate. We are also analysing their relationships with the adjacent tissues of the PrE and trophectoderm, extraembryonic tissues that later participate in the formation of the yolk sac and placenta respectively
Research

Understanding the mechanisms underlying this “developmental programme” is of paramount importance both from a fundamental point of view and for therapeutic applications aimed at using stem cells in regenerative medicine or improving in vitro fertilisation techniques. This programme clearly relies on a complex network of transcription factors and/or repressors as well as intercellular signals. The projects developed in our team are precisely in line with the characterisation of the genetic bases of the regulation of the acquisition of a cellular identity, including pluripotency.
We have demonstrated that the transcription factors NANOG and GATA6 as well as the activation or inhibition of the Fgf signalling pathway are the actors of cell specification. Thanks to the collaboration with the team of G. Dupont (ULB, Brussels) a mathematical model was developed and allowed us to put forward new hypotheses (Frankenberg et al, Dev Cell 2011, Bessonnard et al. Development 2014).
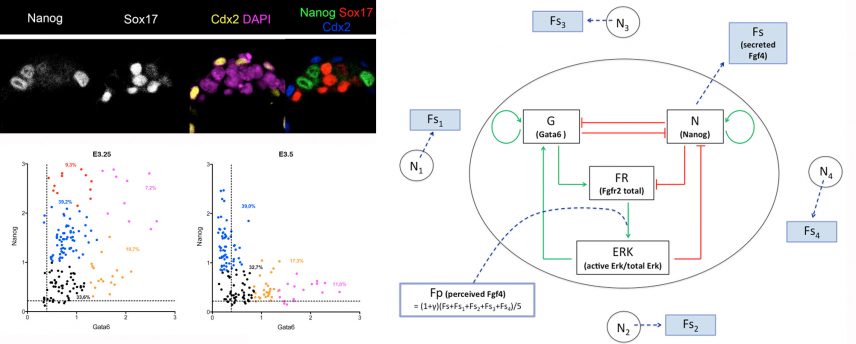
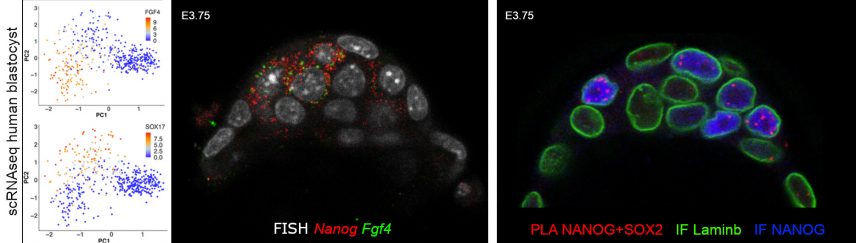
Using transcriptomic analyses on single cells, isolated or in situ within the embryo, we show that Epi cells depend on the coordinated expression of pluripotency markers. An in silico examination of human embryo cells shows that the mechanism of cell differentiation is similar in both species (Allègre et al, Nature Communications 2022).
Proximity Ligation Assay (PLA) detection shows a physical interaction between some of these factors.
Members
Publications
Enhanced Multiplexed Single-Cell RNA-Sequencing for Accurate Detection of Treatment Effects Without Batch Correction in the Avian Embryonic Model
Published on 03 Oct 2025 in Biorxiv
Stephanie Maupetit-Mehouas, Felipe Maurelia Gaete , Pouria Hosseinna , Nicolas Allègre , Yoan Renaud , Jonas Cruzel , Claire Chazaud , Charlène Guillot
Regionally specific levels and patterns of keratin 8 expression in the mouse embryo visceral endoderm emerge upon anterior-posterior axis determination.
Published on 01 Dec 2022 in Frontiers in cell and developmental biology , vol. 10 - pp 1037041
Despin-Guitard E, Quenec'Hdu R , Nahaboo W, Schwarz N, Leube RE, Chazaud C , Migeotte I
NANOG initiates epiblast fate through the coordination of pluripotency genes expression.
Published on 21 Jun 2022 in Nature communications , vol. 13 - pp 3550
Allègre N , Chauveau S , Dennis C , Renaud Y , Meistermann D, Estrella LV, Pouchin P , Cohen-Tannoudji M, David L, Chazaud C
Regulation of the ERK signalling pathway in the developing mouse blastocyst.
Published on 25 Jul 2019 in Development (Cambridge, England) , vol. 146
Azami T, Bassalert C , Allègre N , Valverde Estrella L , Pouchin P , Ema M, Chazaud C
Computational models for the dynamics of early mouse embryogenesis.
Published on 01 Jan 2019 in The International journal of developmental biology , vol. 63 - pp 131-142
Tosenberger A, Gonze D, Chazaud C , Dupont G
Primitive Endoderm Differentiation: From Specification to Epithelialization.
Published on 01 Jan 2018 in Current topics in developmental biology , vol. 128 - pp 81-104
Bassalert C , Valverde-Estrella L, Chazaud C
A multiscale model of early cell lineage specification including cell division.
Published on 09 Jun 2017 in NPJ systems biology and applications , vol. 3 - pp 16
Tosenberger A, Gonze D, Bessonnard S , Cohen-Tannoudji M, Chazaud C , Dupont G
Cripto is essential to capture mouse epiblast stem cell and human embryonic stem cell pluripotency.
Published on 02 Sep 2016 in Nature communications , vol. 7 - pp 12589
Fiorenzano A, Pascale E, D'Aniello C, Acampora D, Bassalert C , Russo F, Andolfi G, Biffoni M, Francescangeli F, Zeuner A, Angelini C, Chazaud C , Patriarca EJ, Fico A, Minchiotti G
Lineage specification in the mouse preimplantation embryo.
Published on 01 Apr 2016 in Development (Cambridge, England) , vol. 143 - pp 1063-74
Chazaud C , Yamanaka Y
Cell Fate Specification Based on Tristability in the Inner Cell Mass of Mouse Blastocysts.
Published on 02 Feb 2016 in Biophysical journal , vol. 110 - pp 710-722
De Mot L, Gonze D, Bessonnard S , Chazaud C , Goldbeter A, Dupont G
Assessment of cell lineages and cell death in blastocysts by immunostaining.
Published on 01 Jan 2015 in Methods in molecular biology (Clifton, N.J.) , vol. 1222 - pp 175-80
Primitive endoderm differentiation: from specification to epithelium formation.
Published on 05 Dec 2014 in Philosophical transactions of the Royal Society of London. Series B, Biological sciences , vol. 369
Gata6, Nanog and Erk signaling control cell fate in the inner cell mass through a tristable regulatory network.
Published on 30 Oct 2014 in Development (Cambridge, England) , vol. 141 - pp 3637-48
Bessonnard S , De Mot L, Gonze D, Barriol M, Dennis C , Goldbeter A, Dupont G, Chazaud C
A close look at the mammalian blastocyst: epiblast and primitive endoderm formation.
Published on 30 Sep 2014 in Cellular and molecular life sciences : CMLS , vol. 71 - pp 3327-38
Artus J, Chazaud C
Fluorescent mRNA labeling through cytoplasmic FISH.
Published on 30 Dec 2013 in Nature protocols , vol. 8 - pp 2538-47
Bmi1 facilitates primitive endoderm formation by stabilizing Gata6 during early mouse development.
Published on 01 Jul 2012 in Genes & development , vol. 26 - pp 1445-58
Lavial F, Bessonnard S , Ohnishi Y, Tsumura A, Chandrashekran A, Fenwick MA, Tomaz RA, Hosokawa H, Nakayama T, Chambers I, Hiiragi T, Chazaud C , Azuara V
Primitive endoderm differentiates via a three-step mechanism involving Nanog and RTK signaling.
Published on 13 Dec 2011 in Developmental cell , vol. 21 - pp 1005-13
Frankenberg S, Gerbe F, Bessonnard S , Belville C , Pouchin P , Bardot O , Chazaud C
The RNA-binding protein Unr prevents mouse embryonic stem cells differentiation toward the primitive endoderm lineage.
Published on 30 Oct 2011 in Stem cells (Dayton, Ohio) , vol. 29 - pp 1504-16
Elatmani H, Dormoy-Raclet V, Dubus P, Dautry F, Chazaud C , Jacquemin-Sablon H
[Early embryogenesis in mammals: stem cells and first commitment steps].
Published on 30 Dec 2008 in Medecine sciences : M/S , vol. 24 - pp 1043-8
Dynamic expression of Lrp2 pathway members reveals progressive epithelial differentiation of primitive endoderm in mouse blastocyst.
Published on 15 Jan 2008 in Developmental biology , vol. 313 - pp 594-602
Gerbe F, Cox B, Rossant J, Chazaud C