Transposable elements (TEs) are mobile genetic sequences that constitute a large fraction of eukaryotic genomes. While their mobility poses a threat to genome integrity, TEs have persisted and expanded, suggesting an evolutionary balance between repression and tolerance. In Drosophila melanogaster, as in other organisms, TE activity is tightly regulated by the piRNA pathway, which ensures transcriptional and post-transcriptional silencing, particularly in gonadal tissues.
Yet, TE repression is not uniformly established during development, and several TEs become transiently expressed in somatic cells. Whether this expression reflects neutral regulatory leakage or functional involvement in developmental processes remains an open question. Some TEs, especially endogenous retroviruses, even encode proteins that may enable intercellular transmission, mimicking viral strategies and challenging the traditional view of TE propagation as strictly cell-autonomous.
Our team investigates the developmental dynamics of TE silencing, the functional potential of TE expression in defined tissues, and the molecular mechanisms enabling soma-to-germline transmission. By studying these processes in Drosophila, we aim to uncover how genomes not only defend themselves against mobile elements but also occasionally engage with them, shaping a complex and evolving relationship between host and TE.
Looking for a PhD, postdoc, or internship opportunity? Get in touch with us.
Funders
Research
Transposable elements (TEs) are abundant and dynamic components of genomes, capable of both disrupting and shaping genetic information. To investigate these dual roles, our team explores how host genomes regulate TEs, how these controls evolve during development, and how some TEs may be repurposed by the host. Using Drosophila melanogaster as a model, we combine genetics, epigenomics, molecular and cellular biology, and developmental approaches to address three main questions:
How is TE silencing established and preserved through development?
We aim to understand how transposable element repression is initiated during development and how this repression is maintained over time in differentiated cells. In this context, using the Drosophila ovarian soma as a model, we combine genetic tools and live imaging to explore the temporal dynamics of piRNA-mediated silencing and to assess the stability and memory of this repression across developmental stages.Exploring the biological significance of TE expression during development
Some TEs escape silencing and are expressed in specific developmental windows or tissues. To better understand these events, we explore their functional consequences, their potential roles in developmental processes, and the host regulatory circuits that allow or restrict them. Through this work, we contribute to redefining TEs not only as threats but also as possible developmental actors.From soma to germline: decoding the infectious potential of endogenous retroviruses
Some retrotransposons, known as endogenous retroviruses (ERVs), retain genes reminiscent of infectious retroviruses, including envelope-like proteins. In particular, in Drosophila, several of these elements are expressed specifically in somatic follicle cells and have the potential to invade the germline. Our objective is to understand the molecular mechanisms that enable such intercellular transfer, with a particular focus on the role of TE-encoded proteins and their interaction with host factors. To this end, we use genetic, molecular, and proteomic approaches to uncover how endogenous retroelements exploit cellular environments to ensure their propagation.
Members
Publications
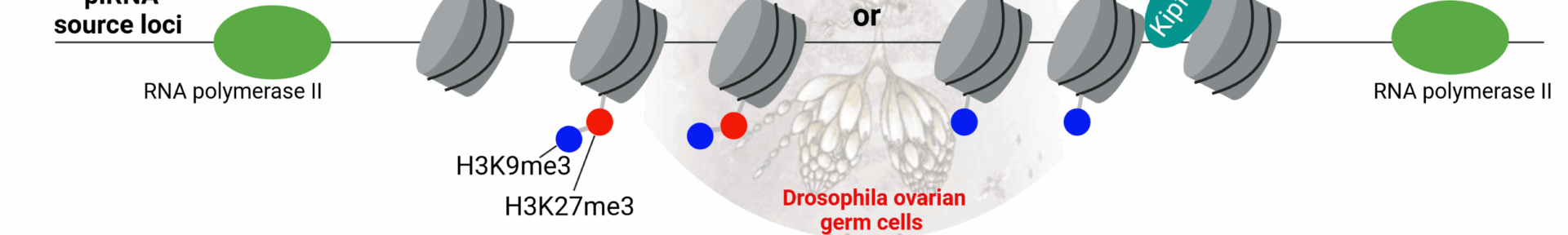
Binding of heterochromatin protein Rhino to a subset of piRNA clusters depends on a combination of two histone marks.
Published on 17 Jun 2025 in Nature structural & molecular biology
Akkouche A , Kneuss E, Bornelöv S, Renaud Y , Eastwood EL, van Lopik J, Gueguen N , Jiang M, Creixell P, Maupetit Mehouas S , Sobieszek A, Gui Y, Czech Nicholson B*, Brasset E* , Hannon GJ*
The transcription factor Traffic jam orchestrates the somatic piRNA pathway in Drosophila ovaries.
Published on 22 Apr 2025 in Cell reports , vol. 44 - pp 115453
Alizada A, Martins A , Mouniée N, Rodriguez Suarez JV, Bertin B , Gueguen N , Mirouse V , Papameletiou AM, Rivera AJ, Lau NC, Akkouche A , Maupetit-Méhouas S, Hannon GJ, Czech Nicholson B, Brasset E
Gap junctions allow transfer of metabolites between germ cells and somatic cells to promote germ cell growth in the Drosophila ovary.
Published on 18 Feb 2025 in PLoS biology , vol. 23 - pp e3003045
Vachias C , Tourlonias C , Grelée L , Gueguen N , Renaud Y , Venugopal P , Richard G , Pouchin P , Brasset E , Mirouse V
The transcription factor Traffic jam orchestrates the somatic piRNA pathway in Drosophila ovaries
Published on 12 Sep 2024 in bioRxiv
Alizada A*, Martins A* , Mouniée N* , Rodriguez Suarez JV*, Bertin B , Gueguen N , Mirouse V , Maupetit-Mehouas S , Rivera AJ, Lau NC, Hannon GJ, Czech Nicholson B, Brasset E.
A dual histone code specifies the binding of heterochromatin protein Rhino to a subset of piRNA source loci.
Published on 11 Jan 2024 in bioRxiv
Akkouche A , Kneuss E, Bornelöv S, Renaud Y , Eastwood EL, van Lopik J, Gueguen N , Jiang M, Creixell P, Maupetit-Mehouas S , Czech Nicholson B, Brasset E , Hannon GJ
Reactivation of a somatic errantivirus and germline invasion in Drosophila ovaries.
Published on 29 Sep 2023 in Nature communications , vol. 14 - pp 6096
Yoth M , Maupetit-Méhouas S , Akkouche A , Gueguen N , Bertin B , Jensen S , Brasset E
The catalytic-dead Pcif1 regulates gene expression and fertility in Drosophila.
Published on 01 May 2023 in RNA (New York, N.Y.) , vol. 29 - pp 609-619
Franco G, Taillebourg E, Delfino E, Homolka D, Gueguen N , Brasset E , Pandey RR, Pillai RS, Fauvarque MO
The histone demethylase Kdm3 prevents auto-immune piRNAs production in Drosophila.
Published on 07 Apr 2023 in Science advances , vol. 9 - pp eade3872
Casier K, Autaa J, Gueguen N , Delmarre V, Marie PP, Ronsseray S, Carré C, Brasset E , Teysset L, Boivin A
More than just an inert dense region.
Published on 14 Oct 2022 in eLife , vol. 11
The Intricate Evolutionary Balance between Transposable Elements and Their Host: Who Will Kick at Goal and Convert the Next Try?
Published on 06 May 2022 in Biology , vol. 11
The Class I HDAC Inhibitor, MS-275, Prevents Oxaliplatin-Induced Chronic Neuropathy and Potentiates Its Antiproliferative Activity in Mice.
Published on 22 Dec 2021 in International journal of molecular sciences , vol. 23
Lamoine S, Cumenal M, Barriere DA, Pereira V, Fereyrolles M, Prival L, Barbier J, Boudieu L, Brasset E , Bertin B , Renaud Y , Miot-Noirault E, Civiale MA, Balayssac D, Aissouni Y, Eschalier A, Busserolles J
Rhino breaks the deadlock in Drosophila testis.
Published on 02 Sep 2021 in PLoS genetics , vol. 17 - pp e1009702
Molla Herman A, Brasset E
Conserved Small Nucleotidic Elements at the Origin of Concerted piRNA Biogenesis from Genes and lncRNAs.
Published on 18 Jun 2020 in Cells , vol. 9
Jensen S , Brasset E , Parey E, Roest Crollius H, Sharakhov IV, Vaury C
Epigenetic Requirements for Triggering Heterochromatinization and Piwi-Interacting RNA Production from Transgenes in the Drosophila Germline.
Published on 10 Apr 2020 in Cells , vol. 9
Komarov PA, Sokolova O, Akulenko N, Brasset E , Jensen S , Kalmykova A
flam piRNA precursors channel from the nucleus to the cytoplasm in a temporally regulated manner along Drosophila oogenesis.
Published on 06 Jul 2019 in Mobile DNA , vol. 10 - pp 28
Environmentally-induced epigenetic conversion of a piRNA cluster.
Published on 15 Mar 2019 in eLife , vol. 8
Casier K, Delmarre V, Gueguen N , Hermant C, Viodé E, Vaury C , Ronsseray S, Brasset E , Teysset L, Boivin A
The interplay between the Argonaute proteins Piwi and Aub within Drosophila germarium is critical for oogenesis, piRNA biogenesis and TE silencing.
Published on 02 Nov 2018 in Nucleic acids research , vol. 46 - pp 10052-10065
Théron E, Maupetit-Mehouas S, Pouchin P , Baudet L , Brasset E , Vaury C
Transcriptional and chromatin changes accompanying de novo formation of transgenic piRNA clusters.
Published on 30 Apr 2018 in RNA (New York, N.Y.) , vol. 24 - pp 574-584
Akulenko N, Ryazansky S, Morgunova V, Komarov PA, Olovnikov I, Vaury C , Jensen S , Kalmykova A
sRNAPipe: a Galaxy-based pipeline for bioinformatic in-depth exploration of small RNAseq data.
Published on 01 Jan 2018 in Mobile DNA , vol. 9 - pp 25
Pogorelcnik R , Vaury C , Pouchin P , Jensen S , Brasset E
Export of piRNA precursors by EJC triggers assembly of cytoplasmic Yb-body in Drosophila.
Published on 08 Dec 2016 in Nature communications , vol. 7 - pp 13739
Increased production of piRNAs from euchromatic clusters and genes in Anopheles gambiae compared with Drosophila melanogaster.
Published on 27 Nov 2015 in Epigenetics & chromatin , vol. 8 - pp 50
George P, Jensen S , Pogorelcnik R , Lee J, Xing Y, Brasset E , Vaury C , Sharakhov IV
PIWI Slicing and RNA Elements in Precursors Instruct Directional Primary piRNA Biogenesis.
Published on 21 Jul 2015 in Cell reports , vol. 12 - pp 418-28
Homolka D, Pandey RR, Goriaux C , Brasset E , Vaury C , Sachidanandam R, Fauvarque MO, Pillai RS
Distinct features of the piRNA pathway in somatic and germ cells: from piRNA cluster transcription to piRNA processing and amplification.
Published on 20 Dec 2014 in Mobile DNA , vol. 5 - pp 28
Microsporidian genomes harbor a diverse array of transposable elements that demonstrate an ancestry of horizontal exchange with metazoans.
Published on 28 Aug 2014 in Genome biology and evolution , vol. 6 - pp 2289-300
Parisot N, Pelin A, Gasc C, Polonais V, Belkorchia A, Panek J, El Alaoui H, Biron DG, Brasset E , Vaury C , Peyret P, Corradi N, Peyretaillade É, Lerat E
History of the discovery of a master locus producing piRNAs: the flamenco/COM locus in Drosophila melanogaster.
Published on 04 Aug 2014 in Frontiers in genetics , vol. 5 - pp 257
Distribution, evolution, and diversity of retrotransposons at the flamenco locus reflect the regulatory properties of piRNA clusters.
Published on 03 Dec 2013 in Proceedings of the National Academy of Sciences of the United States of America , vol. 110 - pp 19842-7
Zanni V, Eymery A, Coiffet M, Zytnicki M, Luyten I, Quesneville H, Vaury C , Jensen S
De novo piRNA cluster formation in the Drosophila germ line triggered by transgenes containing a transcribed transposon fragment.
Published on 30 Jun 2013 in Nucleic acids research , vol. 41 - pp 5757-68
Olovnikov I, Ryazansky S, Shpiz S, Lavrov S, Abramov Y, Vaury C , Jensen S , Kalmykova A
Epigenetics and transgenerational inheritance.
Published on 24 May 2013 in Genome biology , vol. 14 - pp 306
Brasset E , Chambeyron S
NucBase, an easy to use read mapper for small RNAs.
Published on 01 Jan 2013 in Mobile DNA , vol. 4 - pp 1
Transcript levels, alternative splicing and proteolytic cleavage of TFIIIA control 5S rRNA accumulation during Arabidopsis thaliana development.
Published on 30 Jul 2012 in The Plant journal : for cell and molecular biology , vol. 71 - pp 35-44
Layat E , Cotterell S , Vaillant I , Yukawa Y, Tutois S , Tourmente S
Polycomb group-dependent, heterochromatin protein 1-independent, chromatin structures silence retrotransposons in somatic tissues outside ovaries.
Published on 30 Dec 2011 in DNA research : an international journal for rapid publication of reports on genes and genomes , vol. 18 - pp 451-61
An siRNA pathway prevents transgenerational retrotransposition in plants subjected to stress.
Published on 07 Apr 2011 in Nature , vol. 472 - pp 115-9
Ito H, Gaubert H, Bucher E, Mirouze M, Vaillant I , Paszkowski J
Stress-induced activation of heterochromatic transcription.
Published on 28 Oct 2010 in PLoS genetics , vol. 6 - pp e1001175
Tittel-Elmer M, Bucher E, Broger L, Mathieu O , Paszkowski J, Vaillant I
MOM1 and Pol-IV/V interactions regulate the intensity and specificity of transcriptional gene silencing.
Published on 20 Jan 2010 in The EMBO journal , vol. 29 - pp 340-51
Yokthongwattana C, Bucher E, Caikovski M, Vaillant I , Nicolet J, Mittelsten Scheid O, Paszkowski J
The Idefix enhancer-blocking insulator also harbors barrier activity.
Published on 15 Jan 2010 in Gene , vol. 450 - pp 25-31
Genomic environment influences the dynamics of the tirant LTR retrotransposon in Drosophila.
Published on 30 May 2009 in FASEB journal : official publication of the Federation of American Societies for Experimental Biology , vol. 23 - pp 1482-9
Fablet M, Lerat E, Rebollo R, Horard B, Burlet N, Martinez S, Brasset E , Gilson E, Vaury C , Vieira C
Functional characteristics of a highly specific integrase encoded by an LTR-retrotransposon.
Published on 11 Sep 2008 in PloS one , vol. 3 - pp e3185
Faye B, Arnaud F, Peyretaillade E, Brasset E , Dastugue B, Vaury C
Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions.
Published on 12 Jun 2008 in Nature , vol. 453 - pp 948-51
Guelen L, Pagie L, Brasset E , Meuleman W, Faza MB, Talhout W, Eussen BH, de Klein A, Wessels L, de Laat W, van Steensel B
Hypomethylation and hypermethylation of the tandem repetitive 5S rRNA genes in Arabidopsis.
Published on 30 Apr 2008 in The Plant journal : for cell and molecular biology , vol. 54 - pp 299-309
Vaillant I , Tutois S , Jasencakova Z, Douet J, Schubert I, Tourmente S
Maintenance of long-range DNA interactions after inhibition of ongoing RNA polymerase II transcription.
Published on 20 Feb 2008 in PloS one , vol. 3 - pp e1661
Palstra RJ, Simonis M, Klous P, Brasset E , Eijkelkamp B, de Laat W